Learning Objectives
- Explain the roles of ATP, cytoskeletal proteins, and motor proteins in cilia, flagella, and muscle in controlling cell and organism movement
- Differentiate between the different types of muscle (skeletal, smooth, and cardiac)
- Describe the structure of skeletal muscle, and explain the role of this structure in muscle contraction and regulation of muscle contraction through the sliding filament model
- Explain the steps and process of the cross-bridge cycle in muscle contraction
- Describe the structure and function of the neuromuscular junction, and explain the process by which the nervous system and the sarcoplasmic reticulum regulate muscle contraction through availability of calcium
Motor Proteins and Cytoskeletal Tracks
The information below was adapted from Khan Academy: The cytoskeleton. All Khan Academy content is available for free at www.khanacademy.org
The ability to move requires both a firm structure to provide leverage and a mechanism for moving that structure. In multicellular animals, these components are the skeleton and muscles. In single-celled animals and individual cells, these components are typically flagella and/or cilia. Each of these structures rely on both motor proteins and components of the cytoskeleton:
- The cytoskeleton (“cell skeleton”) is a network of filaments that supports the plasma membrane, gives the cell an overall shape, aids in the correct positioning of organelles, provides tracks for the transport of vesicles, and (in many cell types) allows the cell to move. In eukaryotes, there are three types of protein fibers in the cytoskeleton: microfilaments, intermediate filaments, and microtubules.
- Cytoskeletons serve as tracks for movement of motor proteins, which use energy in the form of ATP to “walk along” these cytoskeletal filaments. (This is true of microfilaments and microtubules, but not intermediate filaments which are not associated with motor proteins.)
Microfilaments are also also called actin filaments, because they are composed of actin protein subunits. Microfilaments serve as tracks for the motor protein myosin and are involved in many cellular processes that require motion. In this course, we will focus on their role in muscle cells, where they form organized structures of overlapping filaments called sarcomeres. When the actin and myosin filaments of a sarcomere slide past each other in concert, your muscles contract (much, much more on this later in this page).
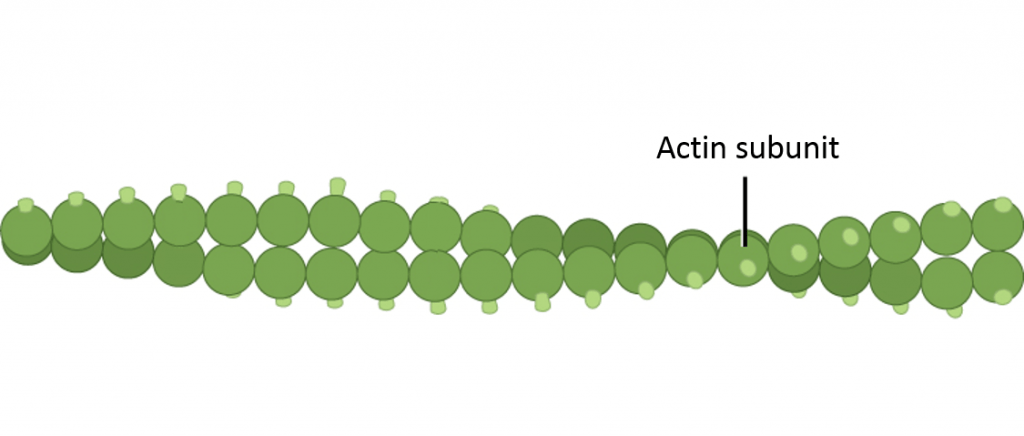
Microtubules are made up of tubulin proteins and serve as tracks for the motor proteins kinesin and dynein; microtubules play important roles in both cellular structural integrity and cell movement. In this course we will focus on their roles in cell movement via cilia and flagella.

Motor proteins use energy in the form of ATP to “walk” along specific cytoskeletal tracks. They are essential for movement of vesicles and other cargoes within cells, as well as for the movement of muscle and cilia/flagella:
- Myosin is associated with actin microfilaments and is required for movement of muscle
- Dynein is associated with tubulin microtubules and is required for movement of cilia and flagella
- Kinesin is associated with tubulin microtubules and is required for movement of vesicles and other intracellular (“within cell”) cargoes
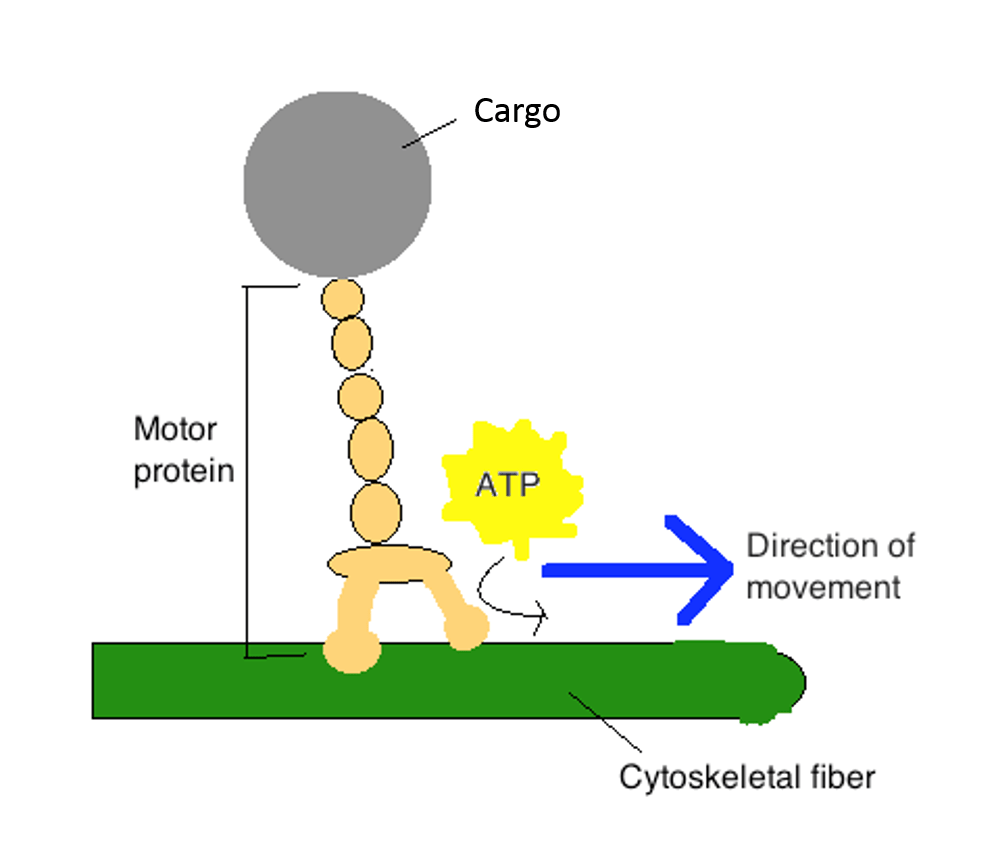
The video below, an excerpt from “The Inner Life of a Cell” by Cellular Visions and Harvard (http://www.studiodaily.com/2006/07/cellular-visions-the-inner-life-of-a-cell/), shows the motor protein kinesin walking along a microtubule track:
Cilia and Flagella
The information below was adapted from Khan Academy: The cytoskeleton. All Khan Academy content is available for free at www.khanacademy.org
ATP, dynein motor proteins, and microtubule tracks are essential for movement of eukaryotic cilia and flagella:
- Flagella (singular, flagellum) are long, hair-like structures that extend from the cell surface and are used to move an entire cell, such as a sperm. If a cell has any flagella, it usually has one or just a few. (Note that, while they carry out the same function, the eukaryotic flagella discussed here has a fundamentally different structure and independent evolutionary origin from the prokaryotic flagella.)
- Cilia (singular, cilium) are similar, but are shorter and usually appear in large numbers on the cell surface. When cells with motile cilia form tissues, the beating helps move materials across the surface of the tissue. For example, the cilia of cells in your upper respiratory system help move dust and particles out towards your nostrils.
Despite their difference in length and number, flagella and motile cilia share a common structural pattern and mechanism driving their movement:
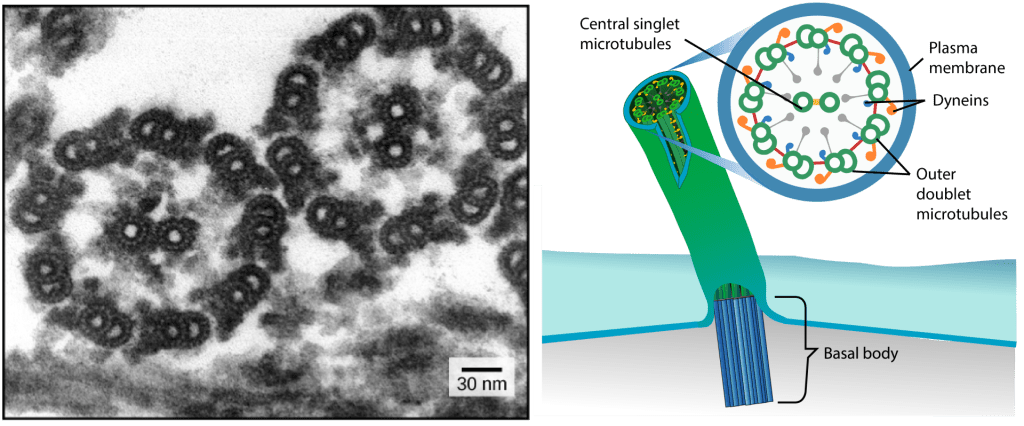
- In most flagella and motile cilia, there are 9 pairs of microtubules arranged in a circle, along with an additional two microtubules in the center of the ring. This arrangement is called a 9 + 2 array. You can see the 9 + 2 array in the electron microscopy image at left, which shows two flagella in cross-section.
- The individual pairs of microtubules are physically connected to each other via protein bridges. They are also physically anchored to the base of the cilium or flagellum.
- The pairs of microtubules are also physically connected via motor proteins called dyneins that move along the microtubules, generating a force that causes the flagellum or cilium to beat. The dynein is anchored to one doublet, and “walks along” the adjacent doublet; the synchronous movement of all dynein molecules across the entire structure causes the cilium or flagellum to bend.
- The structural connections between the microtubule pairs and the coordination of dynein movement allow the activity of the motors to produce a pattern of regular beating, driving the cell to move in a coordinated fashion.
Types of Muscle
The information below was adapted from OpenStax Biology 38.4
ATP, myosin motor proteins, and actin microfilament tracks are essential for contraction of eukaryotic muscle. The vertebrate body contains three types of muscle tissue: skeletal muscle, cardiac muscle, and smooth muscle:
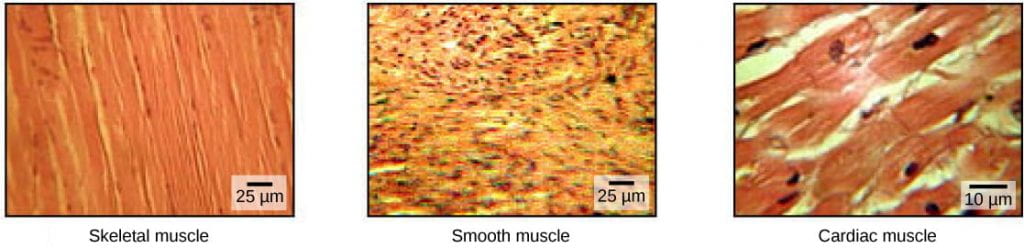
- Skeletal muscles attach to bones or skin and control locomotion and any movement that can be consciously controlled; skeletal muscle is also called voluntary muscle. Skeletal muscles are long and cylindrical in appearance; when viewed under a microscope, skeletal muscle tissue has a striped or striated appearance. The striations are caused by the regular arrangement of contractile proteins (actin and myosin – more on this below).
- Smooth muscle tissue occurs in the walls of hollow organs such as the intestines, stomach, and urinary bladder, and around passages such as the respiratory tract and blood vessels. Smooth muscle has no striations, is under autonomic control; smooth muscle is also called involuntary muscle.
- Cardiac muscle tissue is only found in the heart, and cardiac contractions pump blood throughout the body and maintain blood pressure. Like skeletal muscle, cardiac muscle is striated, but unlike skeletal muscle, cardiac muscle cannot be consciously controlled and is another category of involuntary muscle.
- The nervous system can speed up or slow down the heart rate, but the heart can also beat without nervous system input due to a set of cells called pacemaker cells that spontaneously initiate cardiac muscle contraction.
- Cardiac muscle has intercalated disks, which enable rapid passage of action potentials from one cardiac muscle cell to the next
Skeletal Muscle Structure
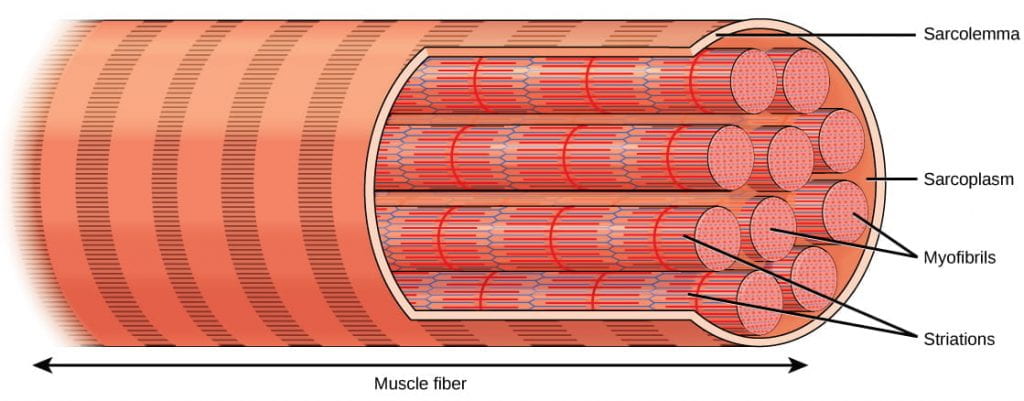
Muscles are composed of structures that enable contraction to promote organismal movement. Each skeletal muscle cell is called a muscle fiber:
- Within each muscle fiber (cell) are myofibrils: long cylindrical structures that lie parallel to the muscle fiber. Myofibrils run the entire length of the muscle fiber, and hundreds to thousands can be found inside one muscle fiber.
- Each myofibril contains repeating units called sarcomeres, which are the actual functional units that cause contraction of a muscle (the “contractile units“). Sarcomeres give muscle its striated or banded appearance, due to the alternating bands of actin and myosin that allow sarcomeres to contract.
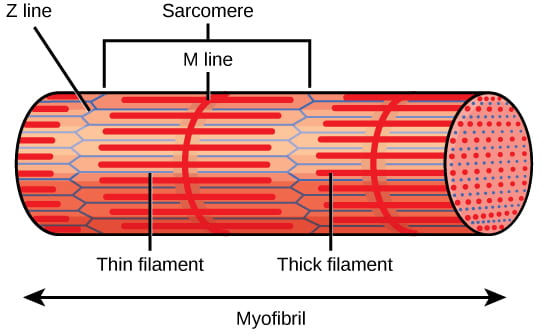
Each sarcomere contains a thick (dark) filament of myosin, and a thin (light) filament of actin:
- The actin filaments are physically anchored to structures at the end of each sarcomere, called Z discs or Z lines.
- The center of the myosin filament is marked by a structure called the M line.
- One sarcomere is the space between two consecutive Z discs.
- A myofibril is composed of many sarcomeres running along its length, and as the sarcomeres individually contract, the myofibrils and muscle cells shorten. Importantly, the actin and myosin fibers stay the same length by sliding past each other as the cell itself shortens.
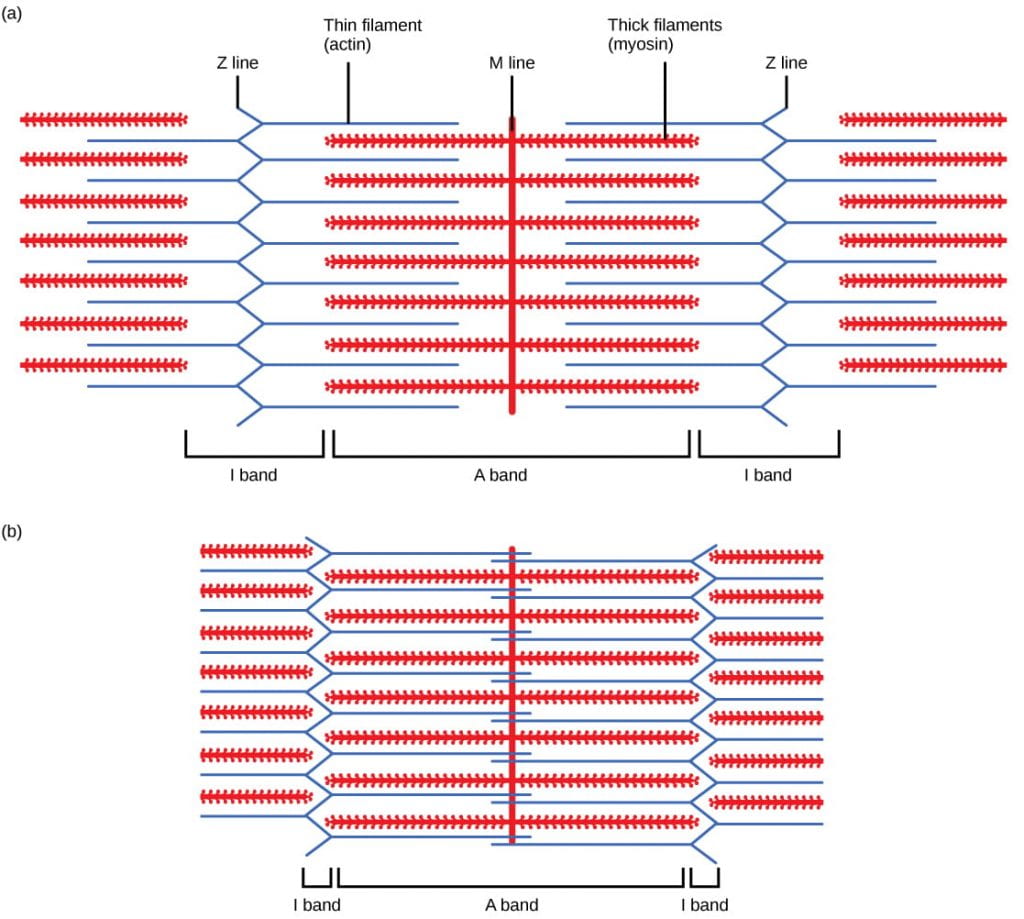
The protein components of the sarcomere include myosin, actin, tropomyosin, and troponin:
- Myosin: Thick filaments are composed of the protein myosin. The tail of a myosin molecule connects with other myosin molecules to form the central region of a thick filament near the M line, whereas the heads align on either side of the thick filament where the thin filaments overlap.
- Actin: The primary component of thin filaments is the actin protein. Actin filaments are attached at the Z disc and extend toward the M line, overlapping with the myosin heads of the thick filament
- Tropomyosin: Actin has binding sites for myosin attachment, but strands of tropomyosin block the binding sites and prevent actin-myosin interactions when the muscles are at rest.
- Troponin: Tropomyosin is regulated by troponin, which is regulated by calcium (Ca2+) ions. The binding of Ca2+ to troponin causes the troponin-tropomyosin complex to move away from the myosin binding sites on the actin filament, allowing myosin to bind to actin to initiate muscle contraction (more on this later in this reading).
The video below describes the organization of muscle fibers:
The Sliding Filament Model of Muscle Contraction
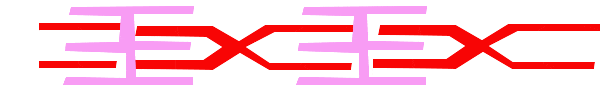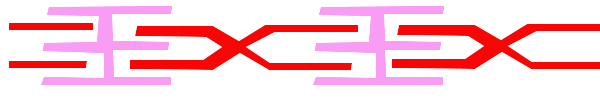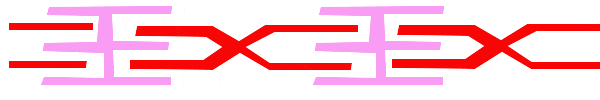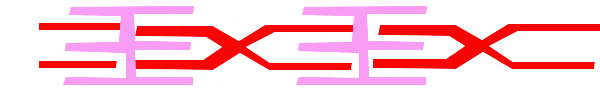
For a muscle cell to contract, the sarcomere must shorten. However, thick and thin filaments (myosin and actin) which comprise the sarcomere do not shorten. Instead, myosin and actin slide by one another, causing the sarcomere to shorten while the filaments remain the same length. The sliding filament model of muscle contraction explains the movement of the bands on the sarcomere at different degrees of muscle contraction and relaxation. The mechanism of contraction is the binding of myosin to actin, forming cross-bridges that generate filament movement. Thus the sliding filament model is accomplished by the cross-bridge cycle of actin-myosin binding (more on the cross-bridge cycle below).
When a sarcomere shortens, the distance between the Z discs is reduced even though the filaments within the sarcomere stay the same length. This is because thin filaments are pulled by the thick filaments toward the center of the sarcomere until the Z discs approach the thick filaments. The filaments appear to slide past each other as illustrated below:
The Cross Bridge Cycle of Muscle Contraction
Muscles contract as myosin heads bind to actin and pull the actin inwards. This action requires energy, which is provided by ATP. Myosin binds to actin at a binding site on the globular actin protein, and myosin also binds to ATP at another binding site, where enzymatic activity hydrolyzes ATP to ADP, releasing an inorganic phosphate molecule and energy. The process works in a cycle (called the “cross bridge cycle”) like this:
- At the start of a cycle, myosin binds to ATP, but it is not yet bound to actin.
- Myosin then hydrolyzes ATP into ADP and inorganic phosphate, and the myosin remains bound to both of these molecules. In this state, it can then attach to a myosin-binding site on the actin thin filament if the binding sites are available (if not, the myosin will remain in this state until the binding sites become available).
- After binding to actin, the myosin protein releases the inorganic phosphate (but stays bound to ADP); the release of inorganic phosphate causes the myosin head to ratchet in what is called “the powerstroke,” pulling the actin thin filament toward the M line and causing muscle contraction.
- After the power stroke is completed, the myosin protein releases ADP. In this state, it remains stuck to the actin filament until it binds another ATP molecule. Because ATP is required for myosin to be able to release the actin, depletion of ATP due to muscle fatigue will cause muscles to remain locked in a contracted state; this is thought to be one of several sources of cramping after exercise.
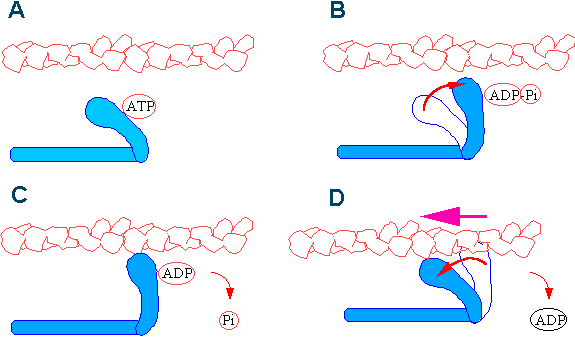
The video below shows the cross bridge cycle of muscle contraction:
Regulation of Muscle Contraction: the Neuromuscular Junction and the Sarcoplasmic Reticulum
Muscle contraction is regulated: there is usually ATP available in the muscle, yet our muscles aren’t permanently contracting. The regulation of muscle contraction is accomplished via the troponin-tropomyosin complex (and the nervous system, which we’ll discuss in detail on the next page). As mentioned above, troponin and tropomyosin determine whether myosin can bind to actin or not:
- Tropomyosin blocks myosin from binding to actin: When a muscle is in a resting state, actin and myosin are separated. To keep myosin from binding to the actin, tropomyosin covers over (blocks) the myosin binding sites on actin molecules, preventing cross-bridge formation and preventing contraction in a muscle without nervous input.
- Tropomyosin is regulated by troponin, and troponin is regulated by calcium: To enable a muscle contraction, tropomyosin must change conformation and uncover the myosin-binding site on an actin molecule and allowing cross-bridge formation.
- Tropomyosin only moves away from actin in the presence of calcium, which is kept at extremely low concentrations in the sarcoplasm (muscle cell cytoplasm). If present, calcium ions bind to troponin, causing conformational changes in troponin that allow tropomyosin to move away from the myosin binding sites on actin.
- Once the tropomyosin is removed, a cross-bridge can form between actin and myosin, triggering contraction. Cross-bridge cycling continues until Ca2+ ions and ATP are no longer available and tropomyosin again covers the binding sites on actin.
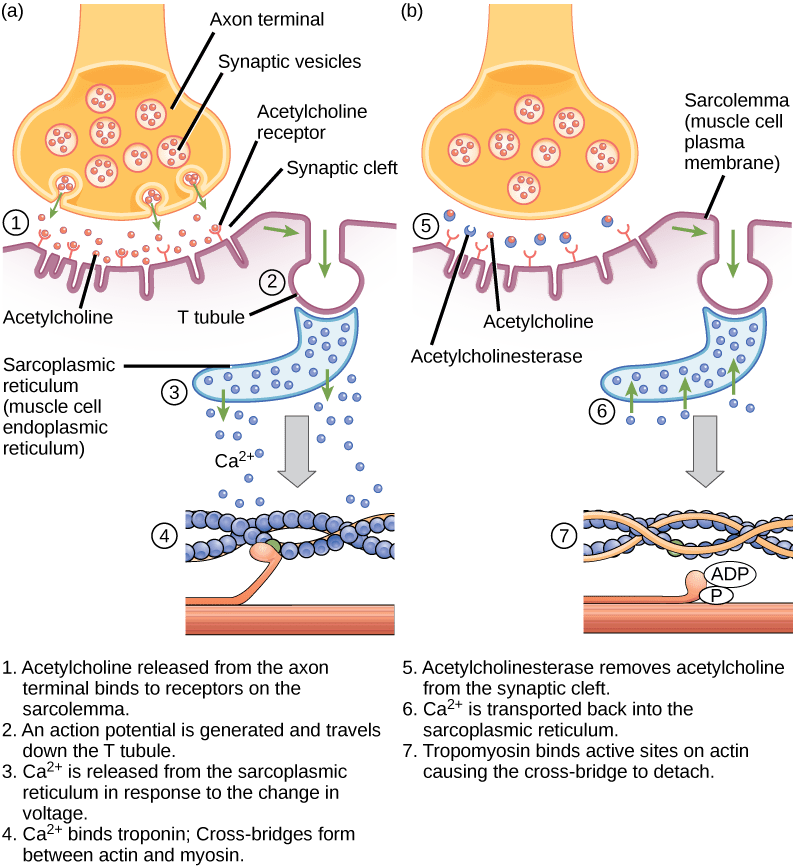
The availability of calcium is regulated by the nervous system. Calcium is stored in a the endoplasmic reticulum, which is called the sarcoplasmic reticulum (SR) in muscle cells. The efferent neurons which control a muscle are synapsed with the muscle in a structure called the neuromuscular junction aka a neuromuscular synapse:
- When an action potential propagates down the axon of the efferent neuron, the neurotransmitter acetylcholine (ACh) is released by the neuron into the synaptic cleft of the neuromuscular junction.
- ACh binds to ACh receptors on the muscle cell, which depolarizes the muscle cell and initiates an action potential in the muscle using exactly the same biochemistry as an action potential in a neuron (influx of sodium ions, efflux of potassium ions)
- The depolarization of the muscle cell then spreads through structures called T-tubules, which carry the action potential into the sarcoplasmic reticulum (SR).
- Depolarization of the SR causes the SR to release calcium into the muscle cytoplasm.
- The sudden presence of calcium in the cytoplasm causes the tropoin-tropomyosin complex to move away from blocking the myosin binding sites on the actin thin filament, allowing muscle contraction to occur.
- After the end of the action potential, the muscle cell returns to its membrane resting potential and calcium is rapidly pumped back into the SR, ending muscle contraction and allowing the muscle to relax (as long as ATP is present to cause myosin to release the actin).
This video describes how neuronal signaling initiates muscle contraction (also called excitation-contraction coupling):
And this video puts it all together to describe how muscle contraction works to facilitate locomotion:

